About Me
A protobiologist and biochemist interested in the emergence of life from complex chemical systems.
With a diverse experimental and computational background, I am passionate about innovation and
discovery through ambitious multidisciplinary research.
I obtained my BSc in Biology from Ben-Gurion University, and my MSc in
Molecular Biology from the Weizmann Institute of Science. I conducted
research into bacterial ecology, immunology, protein evolution and molecular genetics.
Later, I studied the evolution of catalytic chemical networks in the lab of Prof.
Doron Lancet, researching the origin of life in mixed lipid assemblies.
I successfully completed my PhD in Chemistry from the University of Glasgow, under the supervision
of Prof. Lee Cronin. I investigated selection and evolution in chemical and
biological systems, expanding on the Assembly Theory framework, and
developed new analytical tools to uncover causal links at the molecular scale.
My scientific goals include: mapping the informational and functional landscape of living
systems; uncovering the fundamentals of selection and evolution; and generating
self-reproducing protobiological systems in the lab.
When I'm not looking into the origin of life, I enjoy music and podcasts, cooking vegan food,
sports climbing, hiking, going on adventures and continually trying new things.

Research
-
PhD in Chemistry | University of Glasgow
-
2022 - 2025
Selection in Complex Chemical Systems
Researcher in the lab of Prof. Lee Cronin.
Investigated Assembly Theory as a fundamental framework for the study of selection and evolution, through experimentation with advanced analytics and automated systems supported by theoretical work. -
MSc in Molecular Biology | Weizmann Institute of Science
Dean's MSc Prize for Excellence in Research
-
2018-2022
Micellar Origin of Life
Researcher in the lab of Prof. Doron Lancet.
Researched the origin of life in a Lipid World scenario using advanced computational modelling and kinetic simulations of self-reproducing complex catalytic networks. -
2020-2020
Functional Genomics
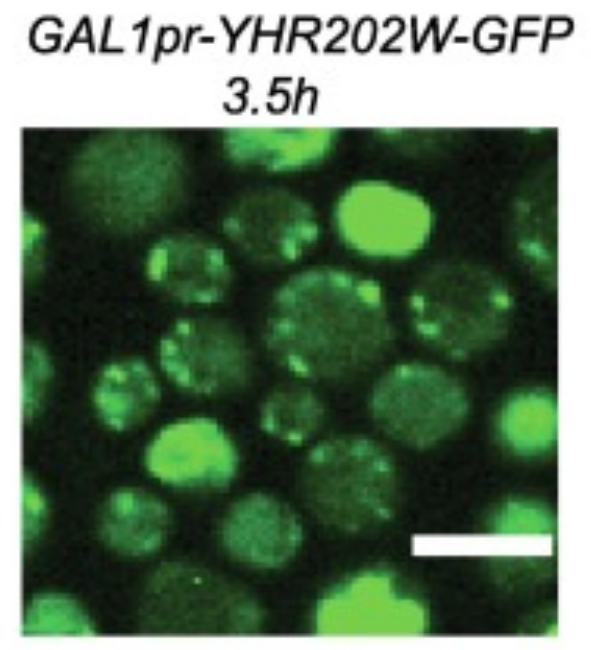
Research Intern in the lab of Prof. Maya Schuldiner.
Predicted the functionality of unexplored enzymes using advanced structural homology, and performed experimental validation. -
2019-2020
Protein Evolution
Research Intern in the lab of Prof. Dan Tawfik.
Examined the functional properties and folding capacities of a primordial RNA-binding protein candidate in the presence of polyamines. -
BSc in Biology | Ben-Gurion University
Ashalim Natural Sciences Honors Program
-
2017-2019
Avian Immunology
Research Assistant in the lab of Prof. Tomer Hertz.
Profiled the antibody repertoires of avian species and identified exposures to influenza and other viruses in relation to migration patterns of wild flocks. -
2018-2019
Microbial Ecology
Research Intern in the lab of Prof. Itzik Mizrahi.
Explored the dynamics of Horizontal Gene Transfer in microbial communities of ruminants.
Publications
Notable Projects:
2024
A Kahana, A MacLeod, H Mehr, A Sharma, E Carrick, M Jirasek, S Walker and L Cronin
arXiv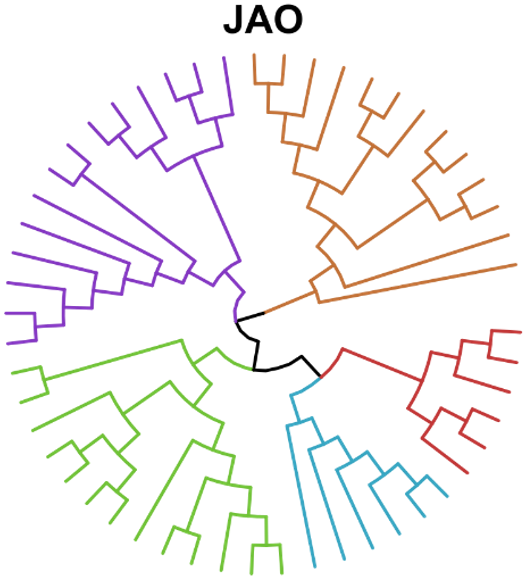
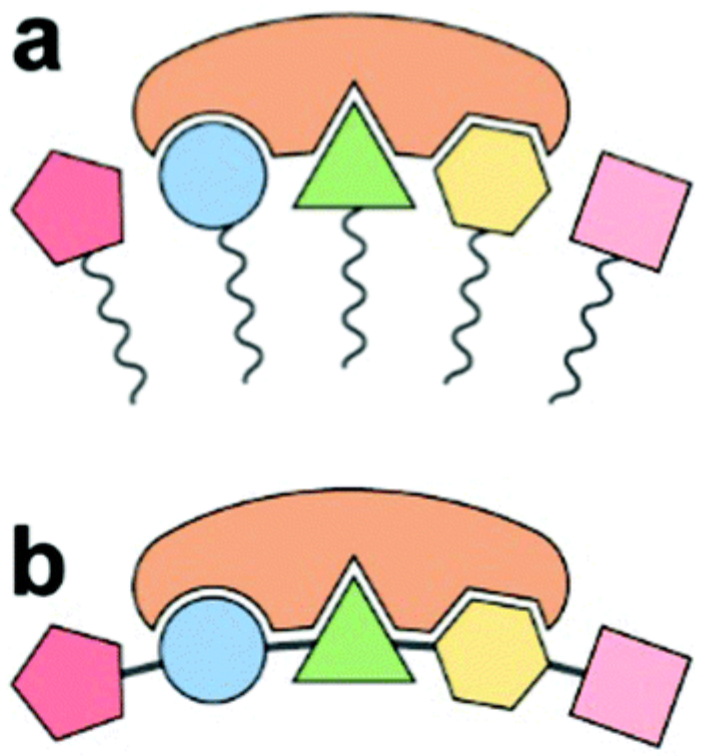
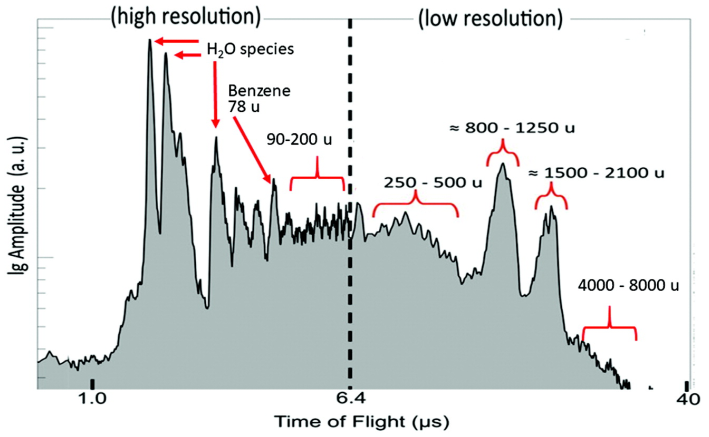
We present a biochemistry-agnostic method to map evolutionary relationships at the molecular scale using mass spectrometry and Assembly Theory, without structure elucidation of analytes. By analyzing diverse biological and non-biological samples, we were able to infer joint assembly spaces of sampled species to determine their biogenicity and taxonomic grouping. We developed an assembly-based phylogenetic tree that aligns with the consensual genome-based model. Later we were able to use our approach to track bacterial lineages exhibiting phenotypic variation. Our results demonstrate we can expand causal molecular inference to non-sequence information without requiring exact molecular identities, thereby opening the possibility to study previously inaccessible biological domains.
2023
A Kahana, L Segev and D Lancet
CellRepPhysSci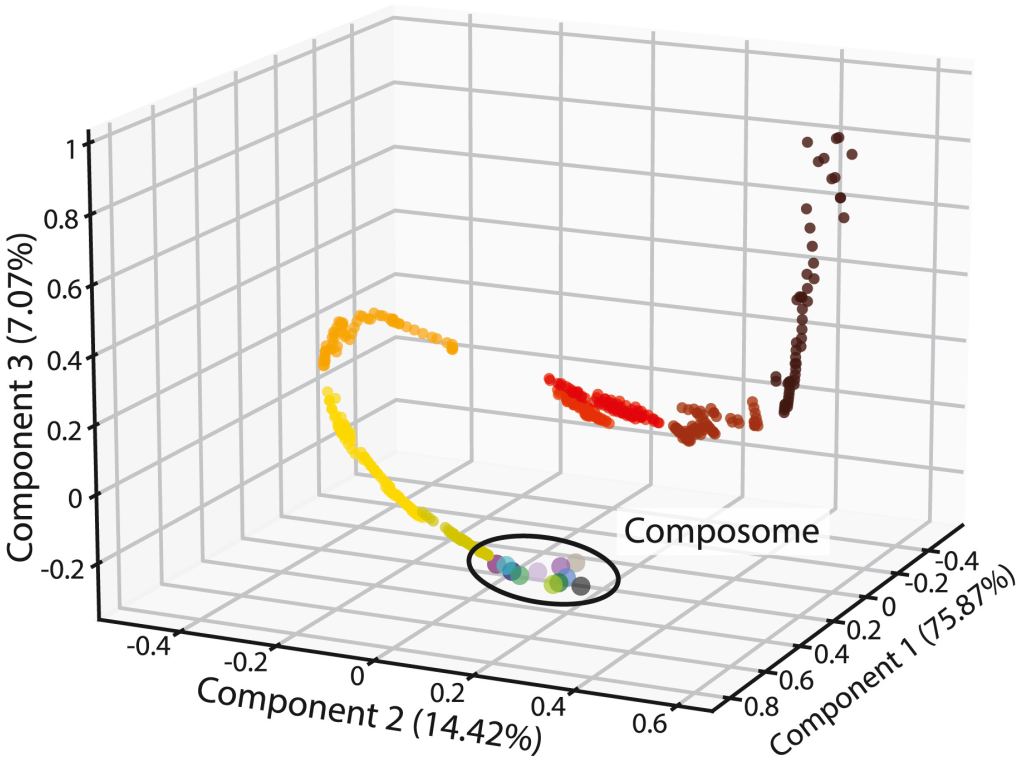
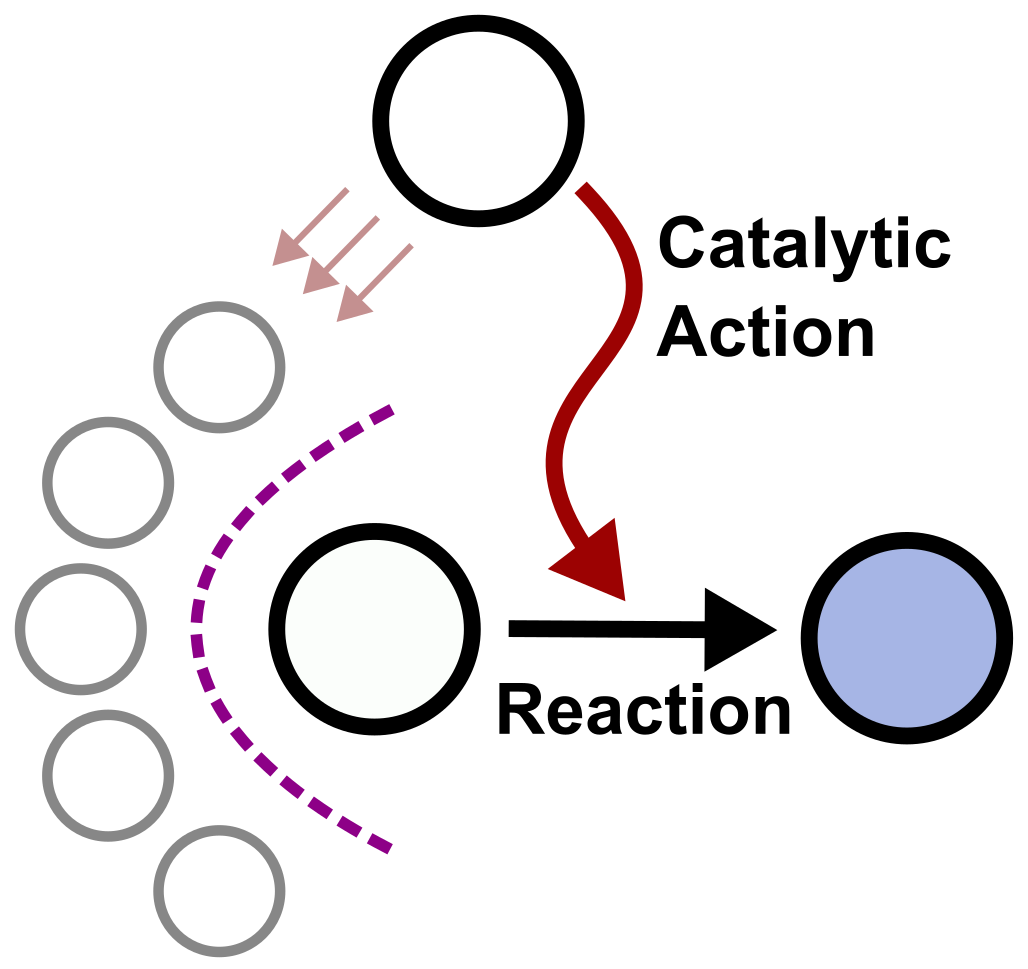
The origin of life likely involved a shift from chaotic chemistry to self-reproducing structures. We examine the reproduction characteristics of the GARD lipid-based model, which simulates physicochemically rigorous, self-replicating networks. The model shows compatibility with heterogeneous environments and portrays trans-generational information transfer. However, we find that self-reproducing states are rare in compositional space, indicating that random exploration would be inefficient to attain reproduction capacities. Rewardingly, we sow thatb all self-reproducing states are dynamic attractors of the catalytic network, suggesting a greatly enhanced propensity for reproduction and primal evolution, augmenting the likelihood of protolife appearance.
2021
A Kahana and D Lancet
NatRevChem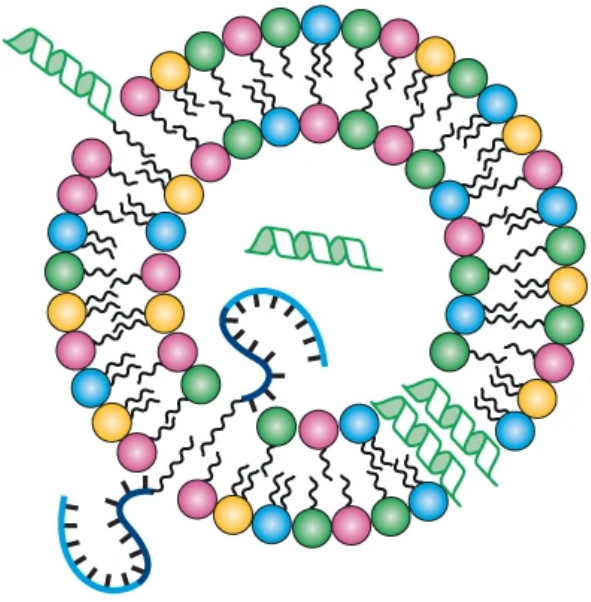
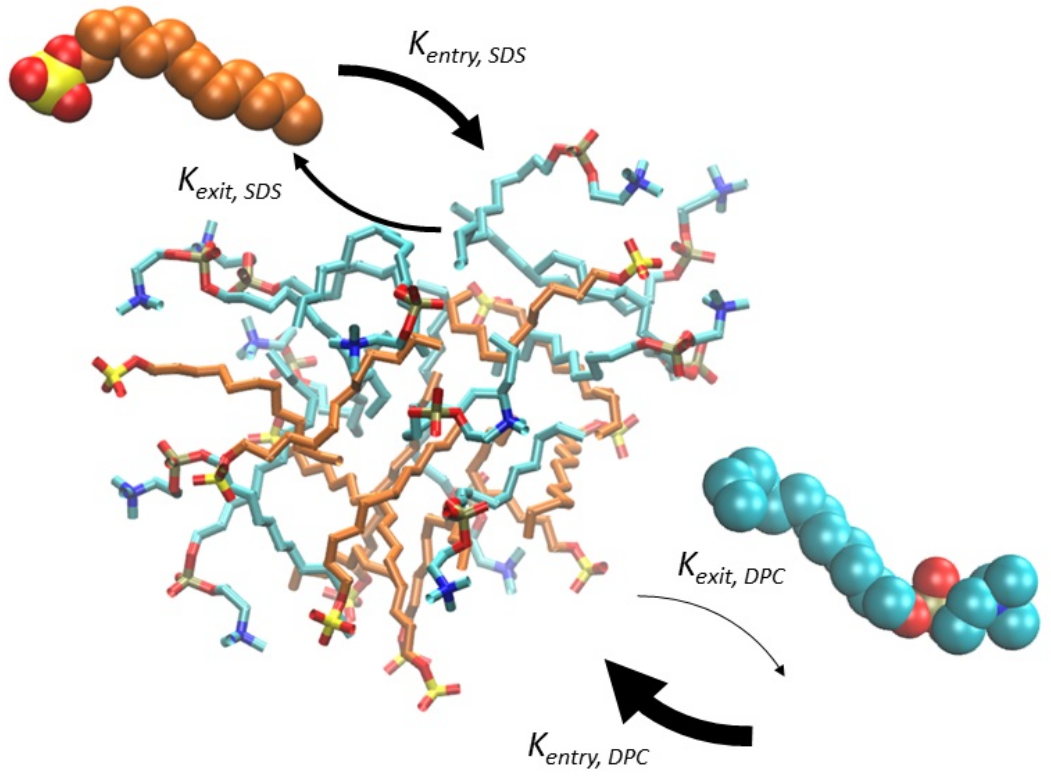
Protocells are often seen as bilayer-enclosed precursors of life, relying on replicating biopolymers for self-reproduction. This Perspective presents an alternative scenario where reproducing lipid micelles with catalytic abilities preceded biopolymer-containing protocells. Support for this idea comes from experiments on micellar catalysis and autocatalytic proliferation, as well as cross-catalysis in mixed micelles leading to life-like dynamics. Our chemically stringent computer-simulated model further shows how catalytic lipid networks could enable micellar compositional reproduction, facilitating primal selection and evolution. Finally, we highlight studies on how endogenously catalysed lipid modifications could guide further protocellular complexification, supporting the idea that lipid micellar assemblies seeded protocellular evolution.
Other Projects:
In Preparation
A Kahana, M Jirasek and L Cronin
2022
A Kahana, D Lancet and Z Palmai
Life2022
A Kahana*, N Cohen* and M Schuldiner
JMB2022
S Levy, M Abd Alhadi, A Azulay, A Kahana, N Bujanover, R Gazit, M A McGargill, L M Friedman and T Hertz
ICB2021
A Kahana, S Maslow and D Lancet
ChemSocRev2020
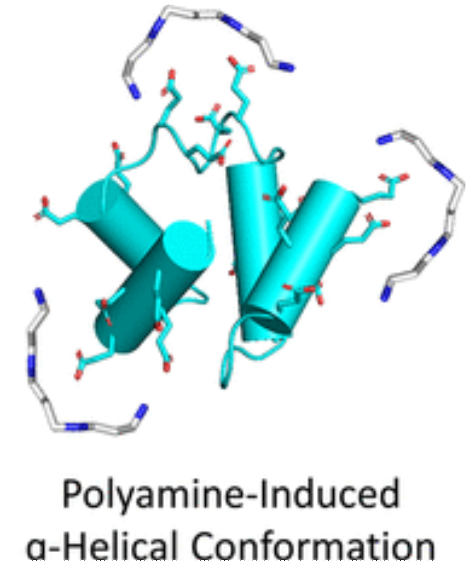
D Despotović, L M Longo, E Aharon, A Kahana, T Scherf, I Gruic-Sovulj and D S Tawfik
Biochemistry2019
A Kahana, P Schmitt-Kopplin and D Lancet
AstrobiologyFor a full list, see my Google Scholar profile.